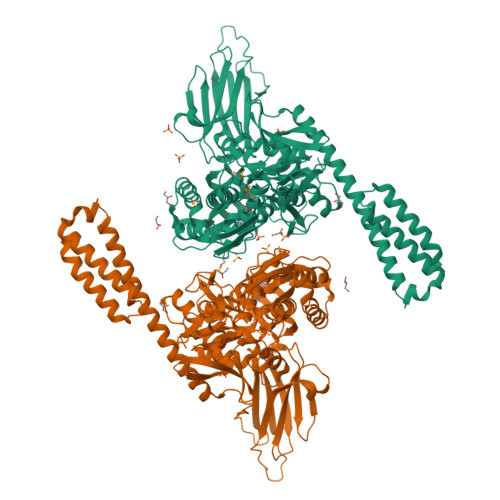
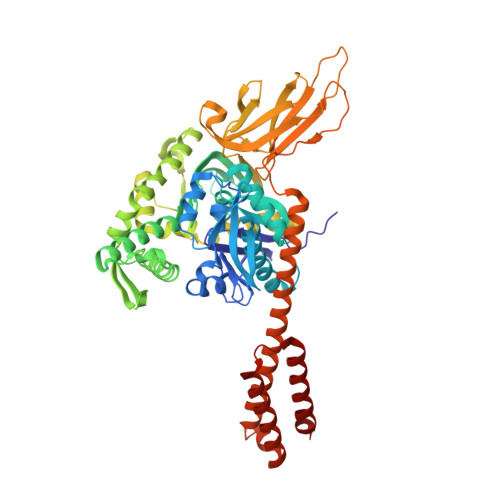
Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s.
Yang, J., Zong, Y., Su, J., Li, H., Zhu, H., Columbus, L., Zhou, L., Liu, Q.(2017) Nat Commun 8: 1201-1201
- PubMed: 29084938
- DOI: https://doi.org/10.1038/s41467-017-01310-z
- Primary Citation of Related Structures:
6ASY - PubMed Abstract:
Cellular protein homeostasis depends on heat shock proteins 70 kDa (Hsp70s), a class of ubiquitous and highly conserved molecular chaperone. Key to the chaperone activity is an ATP-induced allosteric regulation of polypeptide substrate binding and release. To illuminate the molecular mechanism of this allosteric coupling, here we present a novel crystal structure of an intact human BiP, an essential Hsp70 in ER, in an ATP-bound state. Strikingly, the polypeptide-binding pocket is completely closed, seemingly excluding any substrate binding. Our FRET, biochemical and EPR analysis suggests that this fully closed conformation is the major conformation for the ATP-bound state in solution, providing evidence for an active release of bound polypeptide substrates following ATP binding. The Hsp40 co-chaperone converts this fully closed conformation to an open conformation to initiate productive substrate binding. Taken together, this study provided a mechanistic understanding of the dynamic nature of the polypeptide-binding pocket in the Hsp70 chaperone cycle.
Organizational Affiliation:
Department of Physiology and Biophysics, Virginia Commonwealth University, Richmond, VA, 23298, USA.